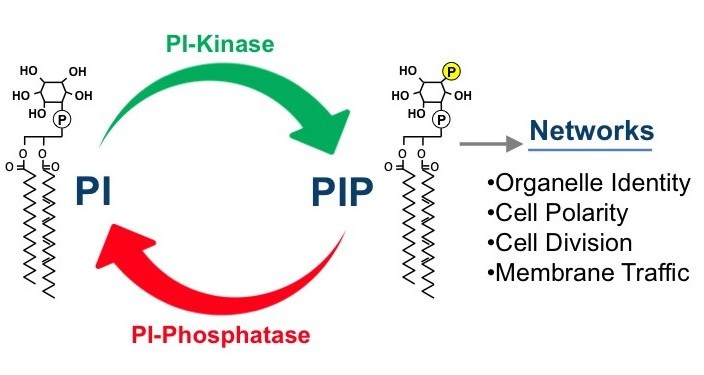
Phosphatidylinositol phosphate (PIP) lipids play an essential role in the regulation of diverse cellular processes, including cell growth, survival, differentiation, cytoskeletal organization, and membrane trafficking. Mutations in PIP metabolic genes are closely linked to serious human diseases including cancer. Specific PIP lipids are enriched in specific subcellular compartments, PI3P is enriched on membranes of the endosomal system, PI3,5P2 on the lysosome, PI4P on the Golgi (and PM) and PI4,5P2 on the plasma membrane. These compartment-specific PIP lipids provide a “lipid identity code” for each subcellular compartment by recruiting and activating specific effector proteins (containing domains such as PH, FYVE, PX, ENTH and GLUE domains) that directly bind the head group of the PI lipids. Our long-term goal has been to understand how the synthesis and turnover of PIPs are temporally and spatially regulated by lipid kinases and phosphatases using the powerful model organism yeast. In addition, we are analyzing the mechanisms by which each PIP regulates its distinct downstream cellular processes through interaction with specific effector proteins.
Synthesis and turnover of each PIP is regulated by a specific set of PIP kinases and phosphatases. Using yeast genetics and molecular biology techniques, and in vitro biochemical techniques, our research has contributed to understanding of the regulatory mechanisms of PIP metabolism. Our recent research includes discovery of unexpected regulatory pathways of PIP metabolism (PI4P and PI4,5P2) and PIP-dependent downstream cellular processes (PI3,5P2). We found that the PI4P phosphatase Sac1 is regulated by the Osh (OSBP) proteins. We also identified the ER-PM contact sites as the key regulatory sites for PI4P metabolism by Sac1. We also have shown that metabolic reprogramming from glycolysis to gluconeogenesis is regulated by PI3,5P2.
Our recent research has focused on the regulation and assembly of the PI 4-kinase and PI4P 5-kinase complexes at the PM, and the identification and function of novel PI3,5P2 effectors.
Key Publications
1. Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, S.D. Emr. (2011). Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 144(3): 389-401.
2. Audhya, A. and S.D. Emr. (2002). Stt4 PtdIns 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev. Cell, : 2: 593-605.
3. Burd, C.G. and S.D. Emr. (1998). Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Molecular Cell. 2(1):157-62.
4. Schu, P. V., K. Takegawa, M. J. Fry, J. H. Stack, M. D. Waterfield and S.D. Emr. (1993). Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science, 260: 88-91.